File formats¶
Input file formats¶
When setting up a CARP simulation, a node, element and fibre files will generally be required. They must be given the same base name, but different extensions, for example:
meshname.pts
meshname.elem
meshname.lon
To support the quick variation of fiber architecture alternative fiber files can be provided using the orthoname option. In this case meshname is used for nodal and element file, but orthoname is used for the fiber file. For instance, using
-meshname biv_rabbit -orthoname biv_rabbit_hf
would read the following files:
biv_rabbit.pts
biv_rabbit.elem
biv_rabbit_hf.lon
Node file¶
Extension: .pts
The node (or points) file starts with a single header line with the number of nodes,
followed by the coordinate of the nodes, one per line, in 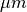 .
For example, for a mesh with
.
For example, for a mesh with 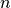 nodes we have:
nodes we have:
| Format | Example |
|---|---|
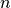 |
10000 |
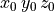 |
10.3400 -15.0023 12.023 |
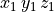 |
11.4608 -12.0237 11.982 |
 |
 |
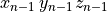 |
-8.4523 36.7472 4.6742 |
Element File¶
Extension: .elem
In general, CARPentry supports various types of elements which can be mixed in a single mesh. However, not all element types are supported in every context, some limitations apply. In electrophysiology simulations element types can be mixed, whereas in mechanics only tetrahedral and hexahedral elements are support. The simple native element file format is as follow
The file begins with a single header line containing the number of elements, followed by one element definition per line. The element definitions are composed by an element type specifier string, the nodes for that element, then optionally an integer specifying the region to which the element belongs.
For example, for a mesh with 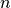 elements and where
elements and where
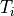 is the type specifier of element
is the type specifier of element 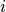 ,
,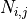 indices the
indices the 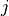 -th node of element
-th node of element 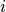 ,
,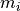 is the number of nodes of element
is the number of nodes of element 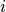 and
and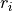 is the optional region of element
is the optional region of element 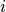 ,
,
the format is given as:
| Format | Example |
|---|---|
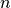 |
20000 |
![T_0 \, N_{0,0} \, N_{0,1} \ldots N_{0,m_i-1} \, [r_0]](_images/math/ad98e8d53e30f7e20f26741a71868844706c94af.png) |
Tt 10 15 123 45 1 |
![T_1 \, N_{1,0} \, N_{1,1} \ldots N_{1,m_i-1} \, [r_1]](_images/math/23595fd5bda2322e1bc352655d76404416808584.png) |
Pr 45 36 100 54 35 74 0 |
 |
 |
![T_{n-1} \, N_{n-1,0} \, N_{n-1,1} \ldots N_{n-1,m_i-1} \, [r_{n-1}]](_images/math/131d78db40ddae8bd3ed8e030c25933f81303dfe.png) |
Tt 9374 7432 1234 4532 1 |
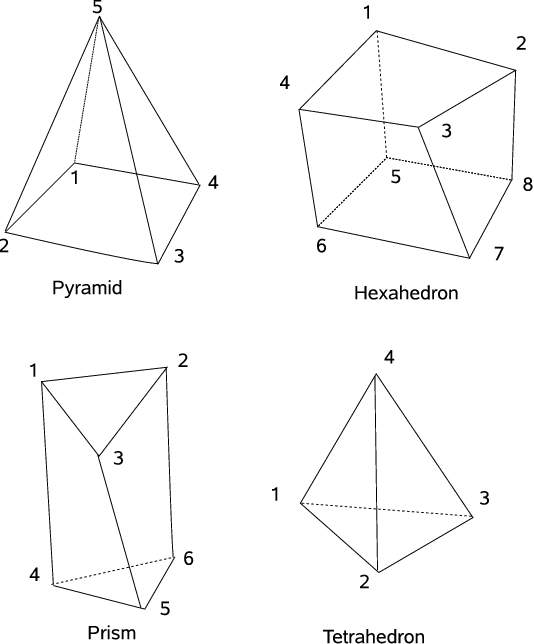
The node indicies are determined by their order in the points file. Note that the nodes are 0-indexed, as indicated in Sec. Node file above. The element formats supported in CARP, including their element type specifier strings are given in table Element types supported in CARPentry for different physics. Electrophysiology (EP), Mechanics (MECH) and Fluid (FL). *: for internal use only, not supported in mesh files.. The ordering of nodes in each of the 3D element types is shown in Nodal ordering of 3D element types.
| Type Specifier | Description | Interpolation | #Nodes | Supported in |
|---|---|---|---|---|
| Ln | Line | linear | 2 | EP |
| cH * | Line | cubic | 2 | HPS |
| Tr | Triangle | linear | 3 | EP |
| Qd | Quadrilateral | Ansatz | 4 | EP |
| Tt | Tetrahedron | linear | 4 | EP, MECH, FL |
| Py | Pyramid | Ansatz | 5 | EP |
| Pr | Prism | Ansatz | 6 | EP |
| Hx | Hexahedron | Ansatz | 8 | EP |
Fiber file¶
Extension: .lon
Fibres are defined in CARP on a per-element basis. They may be defined by just the fibre direction, in which case a traversely isotropic conductivity or mechanics material model must be used, or additionally specifying the sheet (or transverse) direction to allow full orthotropy in the model.
The file format starts with a single header line with the number of fibre vectors defined in the file (1 for fibre direction only, 2 for fibre and sheet directions), and then one line per element with the values of the fibre vector(s). Note that the number of fibres must equal the number of elements read from the element file.
For example, where 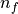 is the number of fibre vectors (1 or 2),
is the number of fibre vectors (1 or 2),
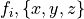 are the components of the fibre vector for element
are the components of the fibre vector for element 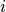 and
and
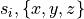 are the components of the sheet vector for element
are the components of the sheet vector for element 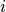 :
:
| Format | Example |
|---|---|
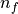 |
2 |
![f_{0,x} \, f_{0,y} f_{0,z} \, [s_{0,x} \, s_{0,y} s_{0,z}]](_images/math/e235747f8fe95364d6fdef8587330baab98027f1.png) |
0.831 0.549 0.077 0.473 -0.775 0.417 |
![f_{1,x} \, f_{1,y} f_{1,z} \, [s_{1,x} \, s_{1,y} s_{1,z}]](_images/math/1d795785895d8ed68ac4fff2b2cde420a01527a3.png) |
0.647 0.026 0.761 -0.678 0.475 0.560 |
 |
 |
![f_{n-1,x} \, f_{n-1,y} \, f_{n-1,z} \, [s_{n-1,x} \, s_{n-1,y} \, s_{n-1,z}]](_images/math/ce1dda8534323b874499f9931a8967ec2be229f7.png) |
0.552 0.193 0.810 -0.806 0.369 0.462 |
The fibre and sheet vectors should be orthogonal and of unit length.
Vertex file¶
Extension: .vtx
An collection of preferably unique mesh indices may be stored in a vertex file.
| Format | Example |
|---|---|
| N # number of nodes | 192 |
| intra|extra # definition domain | intra |
| node_0 | 47 |
| node_1 | 123 |
 |
 |
| node_N-1 | 23943 |
Note
- A stimulus electrode file, e.g. apex.vtx may be added in the parameter file as follows:
-stimulus[0].vtx_file apex
The extra-domain refers to the entire user-specified mesh.
The intra-domain refers to the geometry where non-zero fibers were defined.
Although intra and extra should be the the same for tissue-only geometries, CARPentry/mechanics may not like vtx-files stating the vertices as intra domain.
Purkinje file¶
Extension: .pkje
The format of the Purkinje file is very specific and must be followed carefully.
The # character begins a comment. It and everything following on a line are ignored.
Section purk-file-rules gives the exact rules for defining Purkinje networks.
Below there are comments explaining the different fields inside the file Purkinje.pkje.
Number_of_Cables
Cable 0
Father1 Father2
Son1 Son2
Nodes_Cable
Size
Gap_junction_resistance
Intracellular_conductivity
Node1_x Node1_y Node1_z
...
NodeN_x NodeN_y NodeN_z
Cable 1
...
Note that if a parent or son is unassigned, it is Father2 or Son2 that is assigned a value of -1. An example of a few lines from a Purkinje file follows:
97 # Number of Cables present in the tree [0-96]
Cable 0 # Cable 0 is always the first
-1 -1 # no Fathers for Cable 0
1 2 # Son 1 and Son 2
4 # Number of nodes in this Cable
75 # Relative size of cable (i.e. #parallel fibres or cross-sectional area)
100.0 # gap junction resistance (kOhm)
0.0006 # conductivity (Ohm-cm)
1100 -3400 2200 # NODE 1
1149 -3300 2178 # NODE 2
1184 -3201 2165 # NODE 3
1201 -3090 2154 # NODE 4
Cable 1 # Cable 1, (order must be followed)
0 -1 # FATHER1 = Cable 0, no Father2
3 -1 # SON1 = Cable 3, no Son2
80 # Number of nodes = 80
7.0
100.0
0.0006
1201 -3090 2154
1245 -3020 2143
1209 -2975 2133
.
.
.
Hint
missing link target purk-file-rules
Purkinje-myocardial junction (PMJ) tuning file¶
After the global PMJ parameters are applied, individual junction parameters are set based on values in a file. Cables specified must have a PMJ.
#junctions
cable_no R_PMJ PMJ_scale
cable_no R_PMJ PMJ_scale
.
.
.
cable_no R_PMJ PMJ_scale
Pulse file¶
Note that time values must increase, that is,
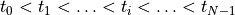
N # Number_of_samples
t0 val0
t1 val1
.
.
.
t(N-1) val(N-1)
Vertex adjustment file¶
N # Number_of_nodes
intra|extra # definition domain
node_0 val_0
node_1 val_1
.
.
.
node_n-1 val_n-1
Neumann boundary file¶
Restitution protocol definition file¶
To be completed Output file formats ——————-
IGB files¶
Extension: .igb
CARP simulations usually export data to a binary format called IGB.
IGB is a format developed at the University of Montreal
and originally used for visualizing regularly spaced data.
An IGB file is composed of a 1024-byte long header followed by binary data
which is terminated by the ASCII form feed character (^L / character 12).
For unstructured grids, the individual dimensions are meaningless
but x 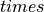 y`
y` 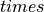
z should be equal to the number of vertices.
The header is composed of strings of the following format,
separated by white space: KeyWord:value
Note that the header must be padded to 1024 bytes.
The first block of key words in IGB keywords need to be specified. The rest are optional.
| KeyWord | Type | Description |
|---|---|---|
| x | int | number of samples in x |
| y | int | number of samples in y |
| z | int | number of samples in z |
| t | int | number of samples in t |
| systeme | string | big_endian or little_endian |
| type | string | binary data type: byte, char, short, long, float, double, int, uint, vec3f, vec4f, vec3d, vec4d |
| unites | string | units of data |
| facteur | float | scaling factor for data |
| zero | float |
|
| org_x | float | lower right x coordinate |
| org_y | float | lower right y coordinate |
| org_z | float | lower right z coordinate |
| org_t | float | time of first slice |
| inc_x | float | distance between x samples |
| inc_y | float | distance between y samples |
| inc_z | float | distance between z samples |
| inc_t | float | time between samples |
| dim_x | float | extent in x |
| dim_y | float | extent in y |
| dim_z | float | extent in z |
| dim_t | float | duration |
| unites_x | string | units of measure for spatial x dimension |
| unites_y | string | units of measure for spatial y dimension |
| unites_z | string | units of measure for spatial z dimension |
| unites_t | string | units of measure for time |
| comment | string | arbitrary comment |
| aut_name | string | author’s name |
| transparent | hex | value for no data |
Dynamic Points¶
Extension: .dynpts
Points may also move in space as functions of time. This is often associated with displacement during contraction, for example. The number of points must remain constant for all time instances, as well as the elements defined by them. Dynamic point files use the IGB format (see IGB files) with data type vec3f.
Vector data files¶
Extension: .vec and .vpts
To display vector data, an auxiliary set of points must be defined by a file with the .vpts suffix. It follows the same format as the Node file. A file with the same base name but having the extension .vec defines the vector and scalar data. It has the following format:
| Format |
|---|
data.x data.y data.z [scalar_datum] |
data.x data.y data.z [scalar_datum] |
 |
data.x data.y data.z [scalar_datum] |
The scalar datum, as indicated, is optional. The .vec format can also be used
for visualization of fiber orientations stored in .lon files.
For this sake, a .vpts file must be generated
holding the coordinates of the centers of each element in the mesh.
This is conveniently achieved with GlElemCenters
and changing the fiber file extension to .vec.
For example, for a mesh with 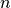 elements
and where
elements
and where 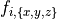 corresponds to the components of the fiber vector
corresponds to the components of the fiber vector 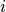 :
:
| Format | Example |
|---|---|
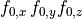 |
8.12453e-01 -2.22623e-01 1.25001e00 |
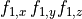 |
5.68533e-01 -1.81807e-01 1.04956e00 |
 |
 |
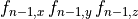 |
5.70671e-01 -1.67572e-01 1.06615e00 |
Vector data can also be input as an IGB file using the types vec3f, vec4f, vec3d, vec4d where 3 or 4 refers to the number of elements in each datum, and d and f refer to float or double. The first 3 elements define the value of the vector field, and the optional 4-th element is the scalar component as above. This file has the suffix .vec.igb.
Auxiliary Grid¶
Extensions: .pts_t, .elem_t, .dat_t
This format is used by Introduction to meshalyzer to display additional data on an auxiliary grid. The grid may contain any of the elements forming the main grid (points,lines,surface elements,volume elements), and the elements may change as a function of time. If scalar data has already been read in, the number of time instances in the auxiliary grid must either be one, or the same as the scalar data. The points file may have only one time instance, which is then assumed constant over all time.
A vertex file is mandatory and has the extension .pts_t. An element file is optional. If present, it has the same base name as the vertex file but with the extension .elem_t. Scalar data may also be optionally defined on the auxiliary grid. The file has the same basename as the vertex file but with the extension .dat_t. The file formats follow:
| .pts_t | .elem_t | .dat_t | ||
|---|---|---|---|---|
| #times | #times | #times | ||
| #points_time0 | #elements_time0 | #data_time0 | ||
| pt(0,0).x | pt(0,0).y | pt(0,0).z | element(0,0) | data(0,0) |
| pt(1,0).x | pt(1,0).y | pt(1,0).z | element(1,0) | data(1,0) |
| . | . | . | . | . |
| . | . | . | . | . |
| . | . | . | . | . |
| . | . | . | element(M,0) | . |
| . | . | . | #elements_time1 | . |
| . | . | . | element(0,1) | . |
| . | . | . | element(1,1) | . |
| . | . | . | . | . |
| pt(N,0).x | pt(N,0).y | pt(N,0).z | . | data(N,0) |
| #points_time1 | . | #data_time1 | ||
| pt(0,1).x | pt(0,1).y | pt(0,1).z | . | data(0,1) |
| pt(1,1).x | pt(1,1).y | pt(1,1).z | . | data(1,1) |
| . | . | . | . | . |
| . | . | . | element(M,1) | . |
| . | . | . | #elements_time2 | . |
| . | . | . | element(0,2) | . |
| . | . | . | . | . |