Single-cell APD restitution
Module: tutorials.01_EP_single_cell.02B_APD_restitution.run
Section author: Jason Bayer <jason.bayer@ihu-liryc.fr>
This tutorial demonstrates how to compute action potential duration (APD) restitution in a single cardiac cell.
Introduction
APD restitution is an important property of cardiac tissue. As pacing frequency is increased, APD shortens to maintain a one to one stimulus to response. For this tutorial, the user will be shown how to construct APD restitution curves to describe the repolarization properties of cardiac cells in response to various pacing protocols. For details see the APD restitution section in the manual.
Problem setup
Single-cell models
This tutorial uses the cardiac cell models in CARP’s LIMPET library and the single-cell tool bench. The single-cell model is a user input. To see a list of all available models, please type the following command.
bench --list-imps
Restitution protocols
Single-cell models are paced with 2-ms-long stimuli at twice capture amplitude with all restitution protocols.
Restitutions protocols are designed to reveal different features of AP restitution.
The S1S2 protocol illustrated in Fig. 56 is used to gauge the propensity
towards developing an arrhythmia.
The dynamic protocol rather reflects restitution for slower transitions between BCLs.
For an S1S2 pacing protocol, the user sets the basic cycle length (BCL) and number of beats, nbeats, for the S1 pacing.
Following S1 pacing, a single S2 beat is applied at a range of coupling intervals (CI) chosen by the user.
The basics of the S1S2 protocol are illustrated in Fig. 56.

Fig. 56 Illustration of the S1-S2 pacing protocol: During an initial pre-pacing period where a number of prebeats stimuli are delivered
at the chosen BCL, the action potential is stabilized.
Alternatively, a snapshot of an AP stabilized at the given BCL can be used and read in using --initial.
After stabilization BCL is kept constant, but prematurity of the CI of the S2 is increased
by decrementing the CI from the starting CI (CI1) (chosen in this experiment to be CI = CI1 = BCL)
with decrements of CIinc down to the final S2 CI of CI0.
When using a dynamic pacing protocol, the S1 cycle length is decremented sequentially and continuously after a user chosen number of beats, nbeats
until the minimum CI of CI0 is reached. The protocol is graphically illustrated in :numref:``fig-apd_restitutions-dynamic-protocol`.
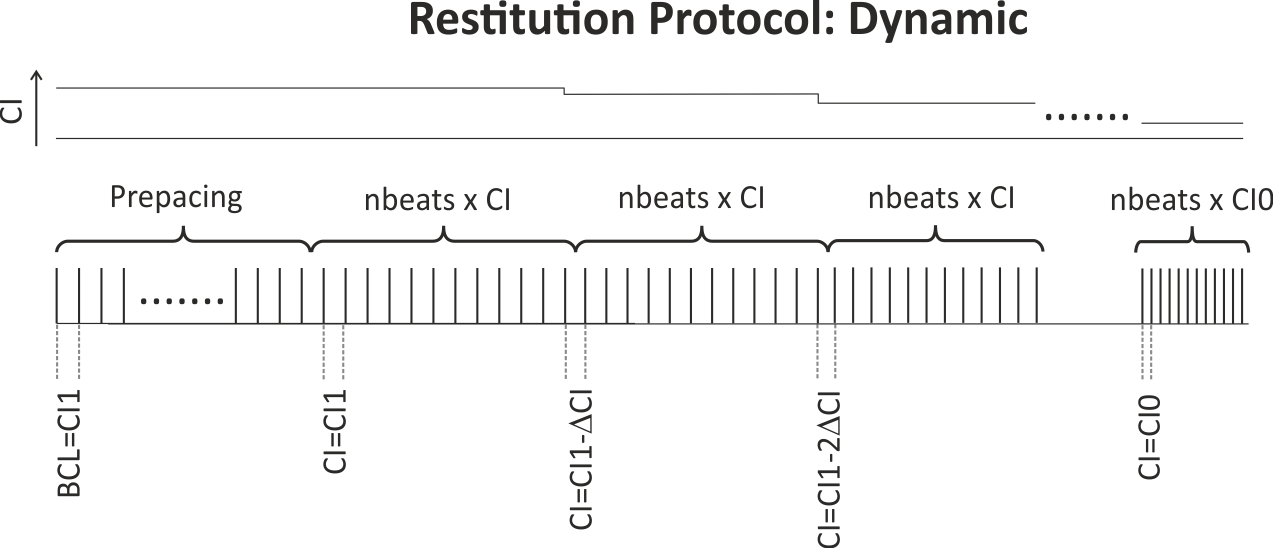
Fig. 57 Illustration of the dynamic restitution protocol: Pacing (again after some prepacing phase or restarting from a stabilized checkpoint for a given BCL)
starts with a train of nbeats stimuli at the longest CI of CI1.
CI is decremented by CIinc after each pulse train until the minimum CI of CI0 is reached.
Action potential duration
Activation potential duration is computed as the time it takes to reach 90% repolarization (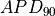 ) from the maximum action potential upstroke potential.
) from the maximum action potential upstroke potential.
Usage
The following optional arguments are available (default values are indicated):
./run.py --help
--Protocol Default: S1S2
Restitution protocol, either S1S2 or Dynamic
--prebeats Default: 20
Number of pre-pacing beats for S1 pacing at BCL
--inital Initial state vector representing limit cycle for chosen BCL
--nbeats Default: 10
Number of beats for S1 pacing at BCL
--CI0 Default: 50 ms
Shortest S2 coupling interval
--CI1 Default: BCL
S1 cycle length and longest S2 coupling interval
--CIinc Default: 25 ms
Decrement for time interval from CI1 to CI0
--imp Default: GTT2_fast
Single-cell model to use
--params Default: ' '
Parameters of single-cell model to modify
The run.py script formats these user inputs into the following file for an S1S2 pacing protocol using bench,
1 # protocol selection 1=S1S2 0=dynamic
prebeats # number of prepacing beats before starting protocol
BCL # basic cycle length
CI1 # S2 prematurity start
CI0 # S2 prematurity end
nbeats # number of beats preceding premature one
CIinc # decrement in S2 prematurity in ms
and the following for a dynamic pacing protocol,
0 # protocol selection 1=S1S2 0=dynamic
prebeats # number of prepacing beats before starting protocol
BCL # initial basic cycle length
CI0 # final basic cycle length
nbeats # number of beats per basic cycle length
CIinc # decrement of bcl in ms
The script run.py then feeds these user input scripts to the function –restitute in bench. The results for DI and APD are output into the ASCII file restout_APD_restitution.dat in the corresponding output directory for the given input parameters. The format of this file is described in the CARP manual under section the describing the function –restitute in bench (3.1.2). The details of the file format are shown below.
Col 1 #Beat : Beat number of an AP
Col 2 Prematurity : P (premature beat) or * (no prematurity)
Col 3 Steady State : O (not in steady state), 1 (in steady state)
Col 4 APD : Action potential duration
Col 5 DI(n) : Diastolic interval of current beat
Col 6 DI(n-1) : Diastolic interval of previous beat
Col 7 Triangulation : APD90-APD30
Col 8 Vm_min : Minimum potential of current beat
Col 9 Vm_max : Maximum potential of current beat
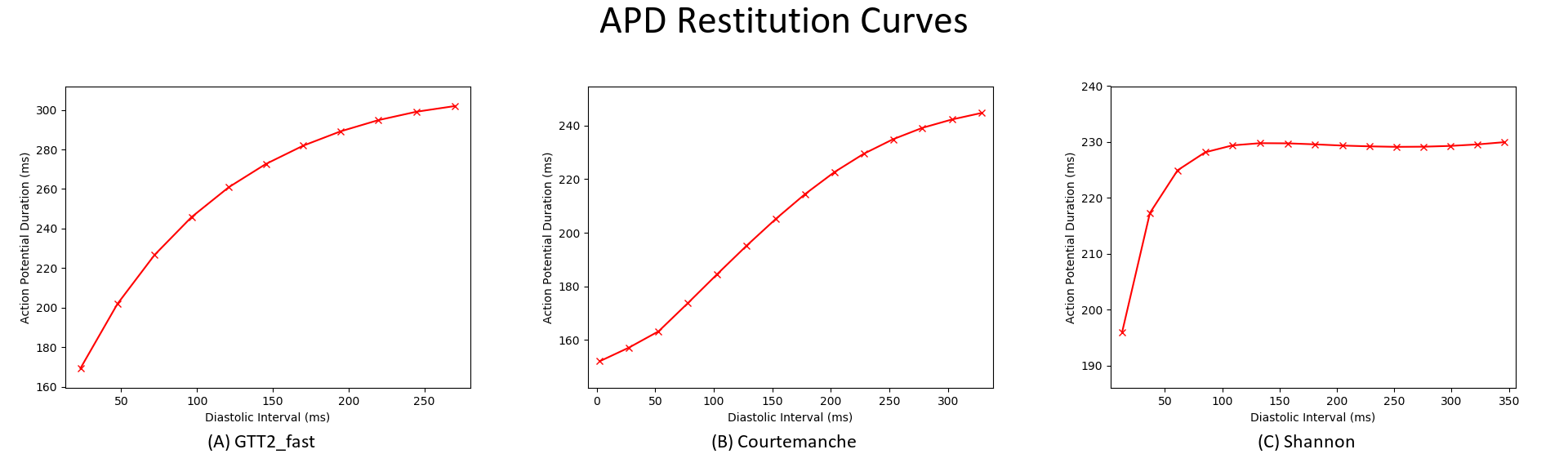
If run.py is ran with the --visualize option, the APD restitution curve (APD Col 4 vs. DI Col 6) in the output file will be plotted using pythons plotting functions.
Note, if the user input for CI0 is too small, you may need to trim the restout_APD_restitution.dat files after a loss of capture occurs. The visualization feature does not remove these values permanently from the output file.
Tasks
To run the experiments of this tutorial change directories as follows:
cd ${TUTORIALS}/01_EP_single_cell/02B_APD_restitution
- Compare the restitution curves for two or more different ionic models. Examples are shown below.
./run.py --imp GTT2_fast --visualize
./run.py --imp COURTEMANCHE --visualize
./run.py --imp SHANNON --visualize
- Using Table 2 from [Tusscher2006], generate a APD restitution curve for the parameter settings -params ‘G_Kr=0.172,G_Ks=0.441,G_pCa=0.8666,G_pK=0.00219,G_tf=2’ and compute the maximum slope of the restitution curve. It should be well above 1.0 and look similar to Figure 5 in the Tusscher2006 paper.
./run.py --imp GTT2_fast --params G_Kr=0.172,G_Ks=0.441,G_pCa=0.8666,G_pK=0.00219,G_tf=2 --visualize
- Do the same as above in 2, but for the parameter settings -params ‘G_Kr=0.134,G_Ks=0.270,G_pCa=0.0619,G_pK=0.0730,G_tf=0.6’. The maximum slope of the restitution curve should be well below 1.0, which promotes alternans and arrhythmogenesis.
./run.py --imp GTT2_fast --params G_Kr=0.134,G_Ks=0.270,G_pCa=0.0619,G_pK=0.0730,G_tf=0.6 --visualize
Notes:
- It could be useful in the future for Gernot to modify the –restitute option to determine CI0 internally rather than having the user determine it.
- An adaptive decrement for S2 would be useful to better define the slope of the APD restitution curve. For example, it is small when APD does not change much to the next decrement, and is increased when APD does change above a certain threshold.
- It would also maybe be useful to print out the Vm for each S1S2 coupling interval to better visualize the restitution behavior. The run.py currently uses the option –res-trace to output restout.txt, but the Vm in the file format is not well described in the carpmanual or bench documentation.
- Lastly, you may need to adjust CI0 and CI1 to the bounds at which the chosen single-cell model was constrained to. The best thing to do is find an APD restitution protocol in the published manuscript for the model, then try to reproduce it.
References
| [Tusscher2006] | ten Tusscher KHWJ, Panfilov AV. Alternans and spiral breakup in a human ventricular tissue model. Am J Physiol Heart Circ Physiol, 291(3):H1088-H1100, 2006. [Pubmed] |